Research Focus
The Transcription Factor p63
| Project Leader: | Prof. Dr. Volker Dötsch (vdoetsch@em.uni-frankfurt.de) |
| Collaborators: | Jakob Gebel, Niklas Gutfreund, Christian Osterburg, Susanne Pitzius, Dr. Frank Löhr, Birgit Schäfer |
The discovery of p63 and of p73, two proteins with high sequence identity to the famous tumor suppressor p53, has sparked speculations that the surveillance of the genetic stability of cells is maintained by an entire network of p53-like tumor suppressors. Surprisingly, however, knock-out mouse studies of all three family members resulted in very different phenotypes. While p53 knock-out mice are mainly characterized by their high frequency of developing tumors at very early age, both the p63 and the p73 knock-out mice show severe developmental defects. Inactivation by mutating the DNA binding domain of p73 results in hippocampal dysgenesis, hydrocephalus, chronic infections and inflammation, as well as abnormalities in pheromone sensory pathways. In addition to these developmental functions of p73, recent analysis of heterozygote knock-out mice (p73+/-) as well as isoform-specific knock-out mouse studies (TAp73-isoform specific knock-out) have suggested that p73 can also act as a tumor suppressor.
The p63 knock-out mice show even more pronounced developmental defects, such as severe limb truncations, lack of a multi-layered skin and other epithelial structures. The p63 knock-out mouse studies led to the identification of mutations in the human p63 gene as the cause of six different human syndromes that are characterized by limb truncations, cleft lip and palates and/or skin abnormalities. The combination of these patient data, the knock-out mice studies and the observation that p63 is highly expressed in the basal layer of stratified epithelial tissues including skin resulted in the model that p63 plays a major role in maintaining epithelial stem cells.
Recently, a second function of p63 was identified: it serves as a quality control factor in female oocytes where p63 is highly expressed. Detection of DNA double strand breaks results in the activation of p63 by phosphorylation and the induction of cell death. In combination with the identification of p63-like genes in nematodes and other invertebrates that are normally not threatened by the development of tumors, this quality control function of germ cells seems to emerge as the ancient function of the entire p53 family. The discovery has also medical relevance: Women treated for cancer with DNA damaging chemo therapy often become infertile. This infertility can now be linked to the activation of p63 which results in the elimination of all oocytes.
Despite the high expression level of p63 in resting oocytes, cell death is not induced without the detection of DNA damage, suggesting that the activity of p63 must be tightly regulated. We have started to characterize the mechanism of the regulation of TAp63α by a combination of biochemical, structural, cell culture and mouse experiments. Our data have revealed that TAp63α in oocytes is kept in a close, transcriptional inhibited and only dimeric conformation. Detection of DNA damage results in the phosphorylation of TAp63α which leads to the opening of the closed dimeric state followed by the formation of active tetramers. This active form has a 20-fold higher DNA binding affinity triggering the expression of PUMA and NOXA which induce apoptosis in the oocytes.
Cell-free Expression & Membrane Proteins
| Project Leader: | Dr. Frank Bernhard (fbern@bpc.uni-frankfurt.de) |
| Collaborators: | Zoe Köck, Roman Levin, Julija Mezhyrova Dr. Frank Löhr, Birgit Schäfer |
Cell-free expression platform for the production of difficult proteins. Cell-free expression is a central technology in the emerging field of Synthetic Biology. Fractionated lysates of special bacterial strains are mixed with a variety of synthetic compounds in order to create tailor made environments for the efficient production and folding of otherwise difficult to obtain proteins and peptides. The high variability and open access of cell-free reactions in combination with efficient and robust bacterial translation pathways provide versatile tools for the production of prokaryotic as well as of eukaryotic proteins. Some unique advantages of cell-free expression are (i) the production of toxic or otherwise inhibitory proteins, (ii) the creation of suitable redox conditions for optimal disulfide bridge formation, (iii) the stabilization of synthesized proteins by addition of co-factors or any other ligand directly into the reaction, (iv) the improved folding of synthesized proteins by using mixed or chaperone enriched cell lysates and (v) the production of even large assemblies by designing reactions with multiple templates or artificial operons. A further particular benefit is the efficient labelling of proteins in any combination as an indispensable prerequisite for structural studies by NMR or crystallization.
The IBPC has established in-house facilities for the production of cell lysates and essential enzymes as well as for a variety of beneficial supplements. Analytical as well as preparative scale cell-free reaction containers for the most efficient two-compartment system have been designed by the IBPC and are manufactured on the campus. Expression screening of new targets, evaluation of new additives as well as the determination of optimal compound concentrations are supported by a robotic pipetting platform operated by custom designed software. Expression reactions are completed within 24 hours and optimized routine protocols yield in between 1-5 mg of target protein per mL or reaction. The IBPC technologies are offered as a flagship platform for cell-free expression in the European Instruct consortium.
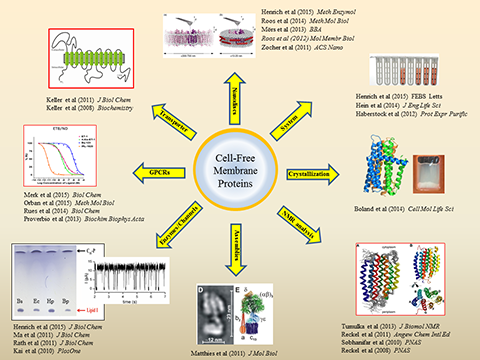
Cell-free expression of membrane proteins. A major research field of the IBPC is the cell-free production of membrane proteins. The system allows to synthesize membrane proteins directly into artificial environments composed out of detergents, other amphiphilic compounds, lipids or any mixtures thereof and thus enables completely new strategies for the folding of hydrophobic proteins. A valuable synergy is the combination of cell-free reactions with pre-formed nano-lipoparticles (nanodiscs) for the co-translational integration of membrane proteins into defined lipid environments. Our published references comprise the characterization of cell-free synthesized transporters, porins, membrane integrated enzymes, ion channels and large assemblies. We have furthermore demonstrated the structural characterization of cell-free synthesized membrane proteins by NMR spectroscopy as well as by crystallization. A current focus of the IBPC is on the characterization of cell-free synthesized G-protein coupled receptors (GPCRs). Efficient production protocols for > 20 different GPCRs have been established and a particular emphasis is on the characterization of human GPCRs involved in hypertension such as the endothelin and vasopressin receptors.
Autophagy
| Project Leader: | Dr. Vladimir Rogov (rogov@bpc.uni-frankfurt.de) |
| Collaborator: | Jessica Huber, Natalja Rogova, Nicole Wesch |
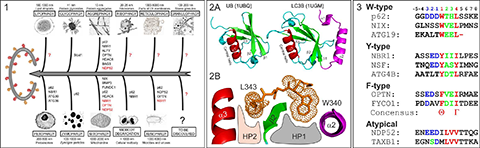
Autophagy sequesters long-living miss-folded and aggregated proteins or/and damaged organelles (cargoes) into autophagosome – double-membrane vesicles, which, in turn, fuse with lysosomes to degrade the enclosed cargoes. The latest studies showed that cells use different types of selective autophagy (e.g., autophagy selective for the degradation of specific types of cargoes (mitophagy – autophagy selective for degradation of mitochondria, aggrephagy – autophagy selective for protein aggregates, etc - Figure 1), as well as kind of nonselective autophagy, where all cellular components near the rapidly growing autophagosome are simply included. Selective autophagy assumes interplay between a few important players, linking the specific nonfunctional cellular component to the degradation machinery: First, the cargo, which frequently gets labeled for degradation by polyUbiquitin-chains. Second, autophagy receptors, which contain specific Ubiquitin-recognition domains (UBA, UBAN, etc). Additionally, the receptors possess a short semi-conserved sequence of amino acids, a so called LIR (LC3 Interaction Region) motif, to recognize the third autophagy players – Ubiquitin-like proteins of ATG8-family, called autophagy modifiers. Being represented in yeast with only one protein (ATG8), these modifiers are present in mammalian cells in several forms – LC3 (LC3A, LC3B and LC3C) and GABARAP (GABARAP, GABARAPL1 and GABARAPL2) related proteins. Similar to Ubiquitin, autophagy modifiers possess a β-sheet wrapped around a central α-helix. However, they have two additional, characteristic α-helices located N-terminally to their UBL core (Figure 2A). This N-terminal subdomain significantly varies among the different members of the LC3/GABARAP family and packs onto the UBL core to form a deep hydrophobic pocket (designated HP1). Another hydrophobic pocket (HP2) is formed by the hydrophobic residues of the central α-helix and β-strand 2 of the UBL core (Figure 2B). The two hydrophobic pockets and β-strand 2 of LC3/GABARAPs form a structural platform for recognition of various LIR motifs (Figure 3), engaging in interaction networks associated with autophagy and membrane trafficking. All three players – Ubiquitin, autophagy receptors and autophagy modifiers – are connected to each other by specific networks of protein-protein interactions (for review, see Mizushima et al., 2011; Rogov et al., 2014). These interactions are precisely regulated to provide a fast and specific answer to various cellular stresses.
In our institute we combine variety of structural (high-resolution NMR spectroscopy and X-ray analysis), biophysical (CD, ITC and SPR) and biochemical methods to understand basic molecular principles of selective autophagy on each it step: recognition of the marked for degradation cargoes by autophagy receptors, recognition of the charged autophagy receptors by autophagy modifiers, specificity and selectivity of the interactions between various LIRs (natural and artificial) and the autophagy modifiers, etc. We are investigating regulation of these specific autophagy interactions by post-translational modifications, chemical components, cellular compartmentalization, mutagenesis, etc. We are looking for physical connections between several key signaling pathways - ubiquitination, ufmylation and others - to selective autophagy.
The Institute of biophysical chemistry is a part of the several research programs aimed to investigate the autophagy and use it in therapeutic practice (SFB 1177, DKTK).
Figures:
Selected publications:
| Nix is a selective autophagy receptor for mitochondrial clearance. Novak I, Kirkin V, McEwan DG, Zhang J, Wild P, Rozenknop A, Rogov V, Löhr F, Popovic D, Occhipinti A, Reichert AS, Terzic J, Dötsch V, Ney PA, Dikic I. EMBO Rep. 2010 Jan;11(1):45-51. doi: 10.1038/embor.2009.256. Epub 2009 Dec 11. |
||
| Phosphorylation of the autophagy receptor optineurin restricts Salmonella growth. Wild P, Farhan H, McEwan DG, Wagner S, Rogov VV, Brady NR, Richter B, Korac J, Waidmann O, Choudhary C, Dötsch V, Bumann D, Dikic I. Science. 2011 Jul 8;333(6039):228-33. doi: 10.1126/science.1205405. Epub 2011 May 26. |
||
| Characterization of the interaction of GABARAPL-1 with the LIR motif of NBR1. Rozenknop A, Rogov VV, Rogova NY, Löhr F, Güntert P, Dikic I, Dötsch V. J Mol Biol. 2011 Jul 15;410(3):477-87. doi: 10.1016/j.jmb.2011.05.003. Epub 2011 May 18. |
||
| Structural basis for phosphorylation-triggered autophagic clearance of Salmonella. Rogov VV, Suzuki H, Fiskin E, Wild P, Kniss A, Rozenknop A, Kato R, Kawasaki M, McEwan DG, Löhr F, Güntert P, Dikic I, Wakatsuki S, Dötsch V. Biochem J. 2013 Sep 15;454(3):459-66. |
||
| Interactions between autophagy receptors and ubiquitin-like proteins form the molecular basis for selective autophagy. Rogov V, Dötsch V, Johansen T, Kirkin V. Mol Cell. 2014 Jan 23;53(2):167-78. doi: 10.1016/j.molcel.2013.12.014. Review. |
||
| CUL3-KBTBD6/KBTBD7 ubiquitin ligase cooperates with GABARAP proteins to spatially restrict TIAM1-RAC1 signaling. Genau HM, Huber J, Baschieri F, Akutsu M, Dötsch V, Farhan H, Rogov V, Behrends C. Mol Cell. 2015 Mar 19;57(6):995-1010. doi: 10.1016/j.molcel.2014.12.040. Epub 2015 Feb 12. |
||
| TECPR2 Cooperates with LC3C to Regulate COPII-Dependent ER Export. Stadel D, Millarte V, Tillmann KD, Huber J, Tamin-Yecheskel BC, Akutsu M, Demishtein A, Ben-Zeev B, Anikster Y, Perez F, Dötsch V, Elazar Z, Rogov V, Farhan H, Behrends C. Mol Cell. 2015 Oct 1;60(1):89-104. doi: 10.1016/j.molcel.2015.09.010. |




